In 2016 three of us (Ola Hössjer, Ann Gauger and Colin Reeves) published an algorithm to simulate population genetics scenarios in a more flexible and detailed way than has been done before, using a backwards simulation method. This algorithm has now been implemented in a high-performance C++ code. This is the algorithm described in the paper A Single-Couple Human Origin is Possible, which was used for the bulk of the analysis, including all the work involving linkage disequilibrium. The method of this algorithm greatly increases its computing power–the depth of sequence estimations that are possible and the speed at which computations can be made are greatly increased.

Here we explain some of why the backwards simulation method is so efficient and allows much larger detailed simulations than were previously possible.
Forward Simulation
The most obvious way to simulate the genetic evolution of a population is simply to create some imaginary ancestors, and then simulate the process of reproduction, recombination and mutation down the generations.
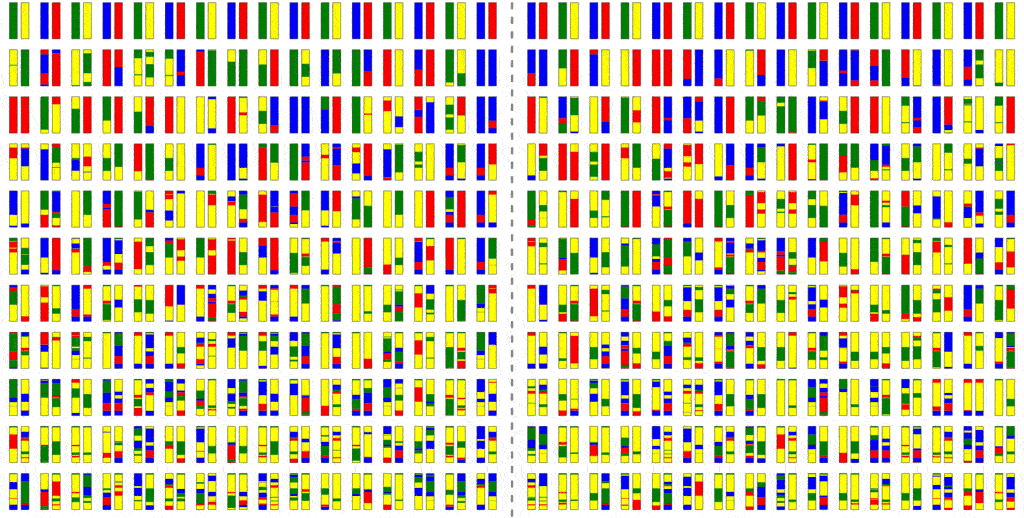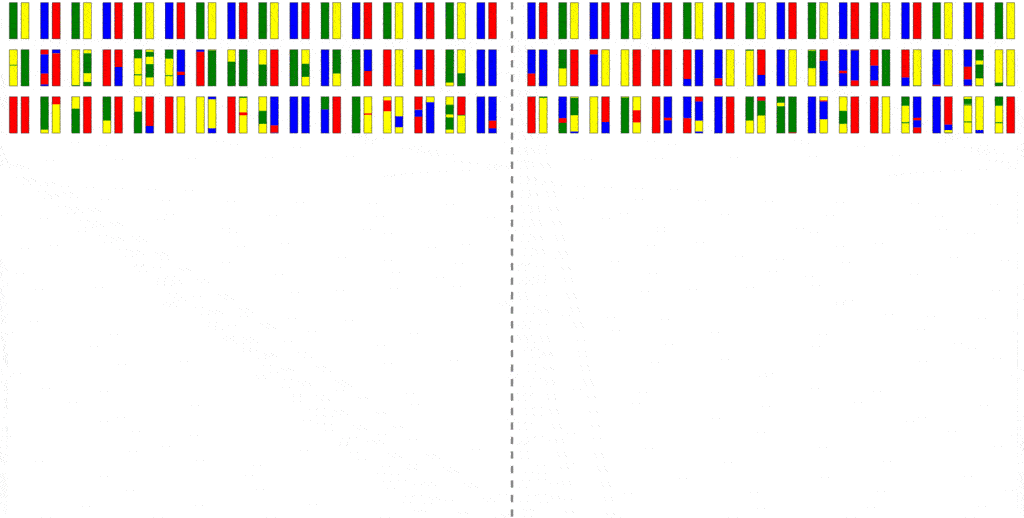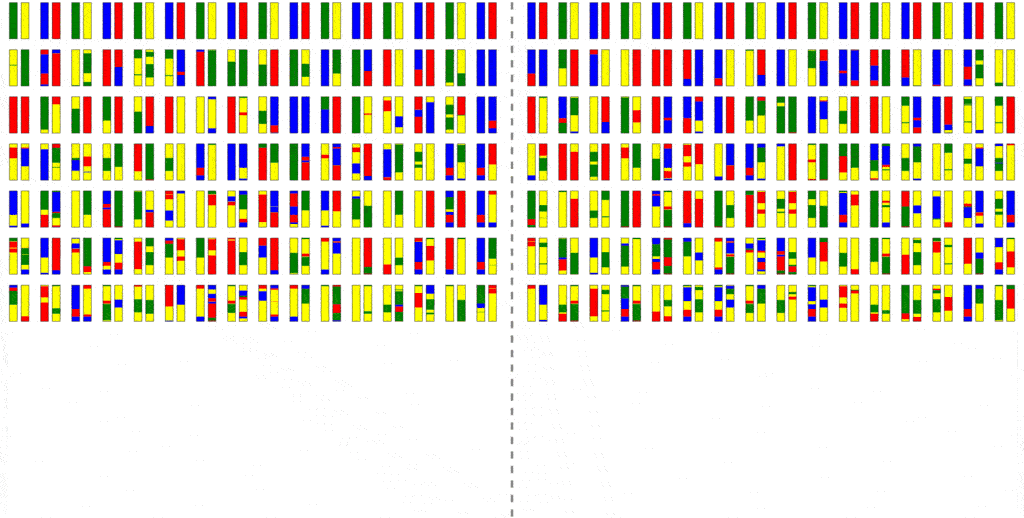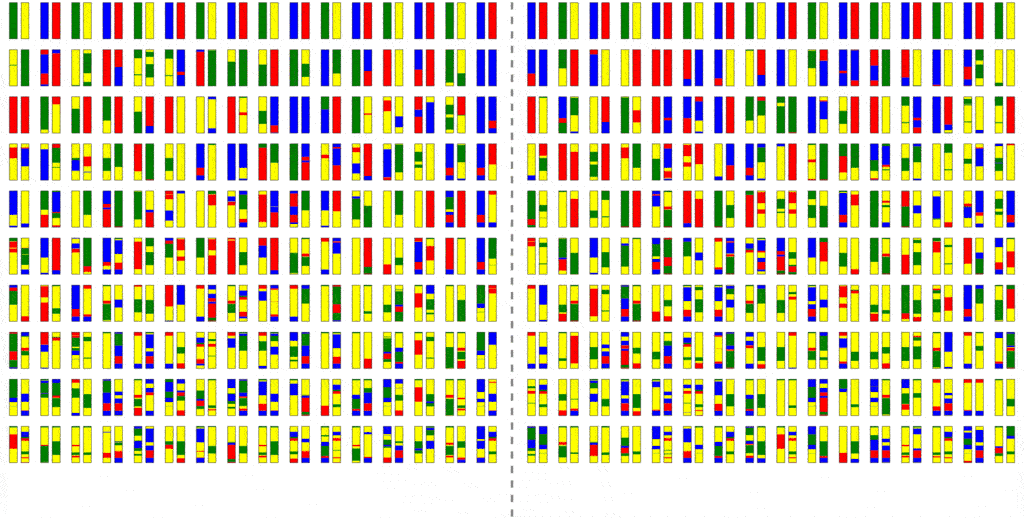
However, as we saw in basic population genetics, many of these chromosomes are non-ancestral, and the computing-power put into calculating them is wasted.
Backwards Simulation – Individuals
This can be improved a little bit by simulating into two stages: First we trace the relationships between people, but backwards in time, in order to avoid computing those that are non-ancestral. Then we generate the truly ancestral members of the initial population, and simulate the mutation process forward.
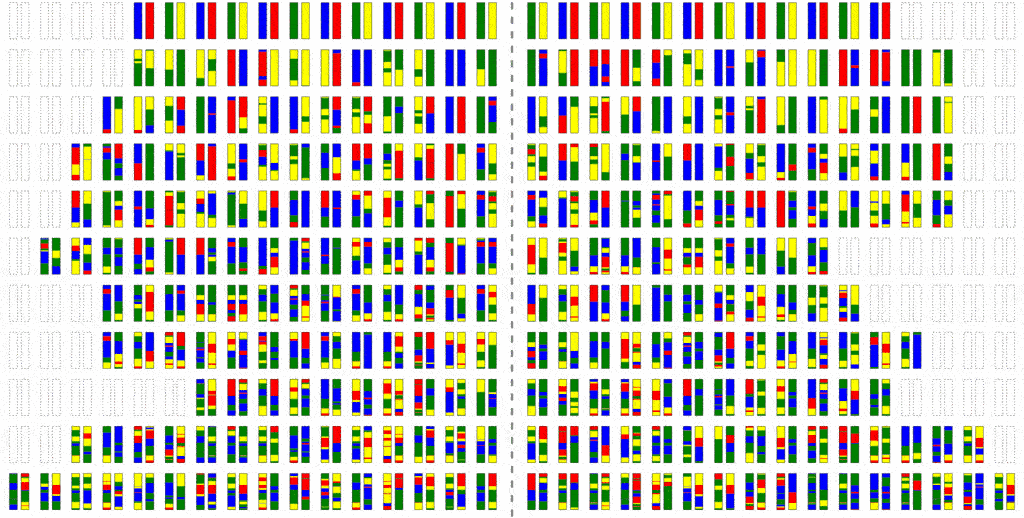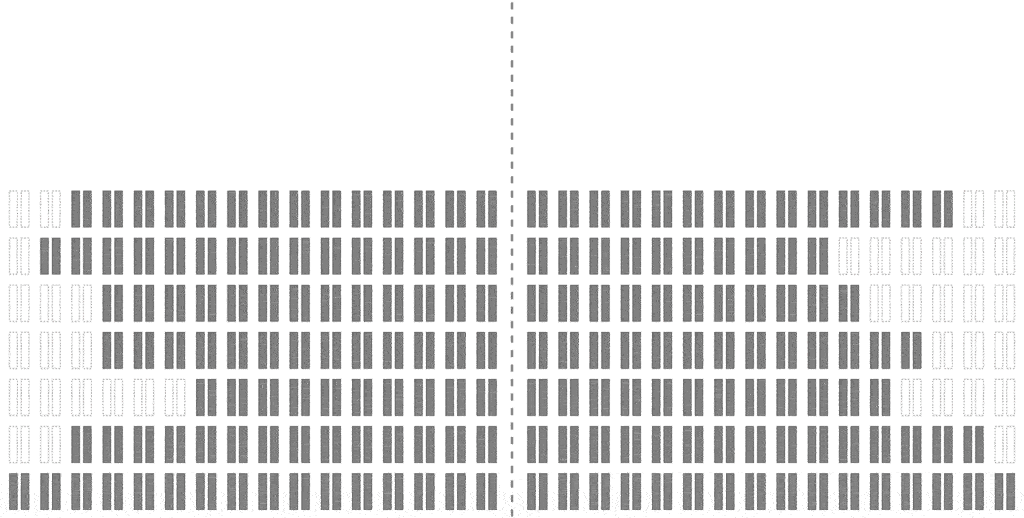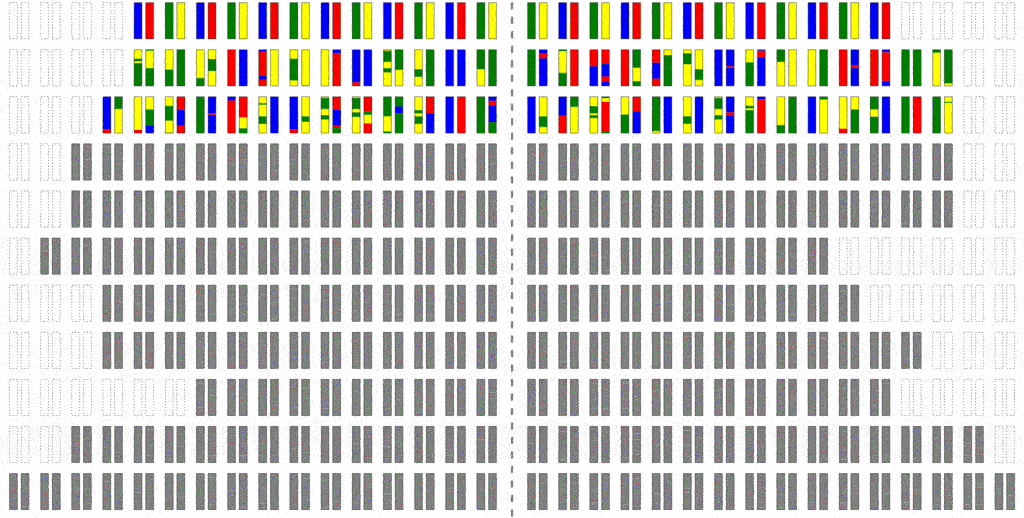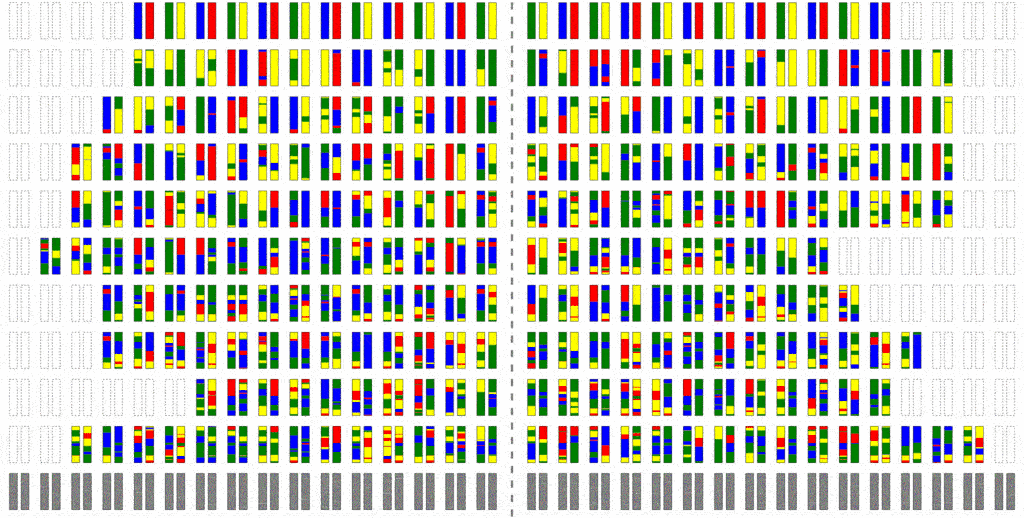
This saves a little bit of computation, but not all that much; the problem is that because where there is recombination, most members of a past population will be ancestors. To see why intuitively, consider the fact that you have two parents, four grandparents, eight great-grandparents and so on. It doesn’t take very many generations before your ancestors include most of the human race. It is this use of haploblocks that gave Haplo its name.
Backwards Simulation – Haploblocks
The efficiency of the computation can be vastly increased if we trace the relationships between haplotypes instead of just individuals, by simulating parental relationships and recombination patterns, backwards from the present, and then propagating the remaining haplotypes forward, adding mutations.
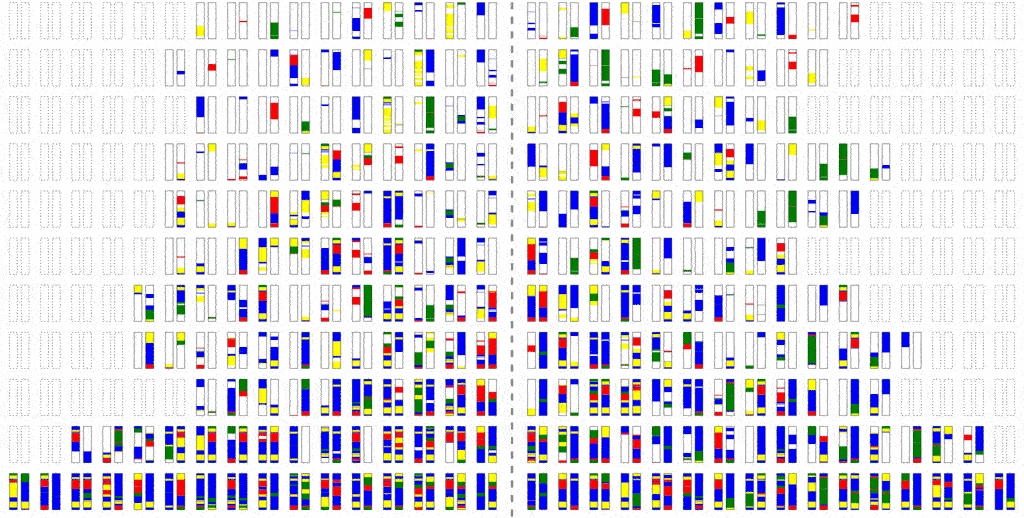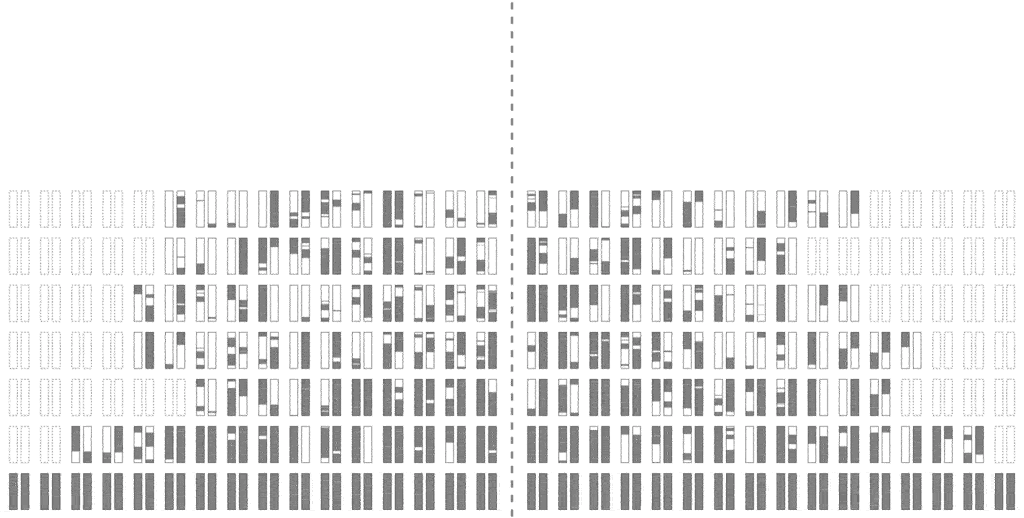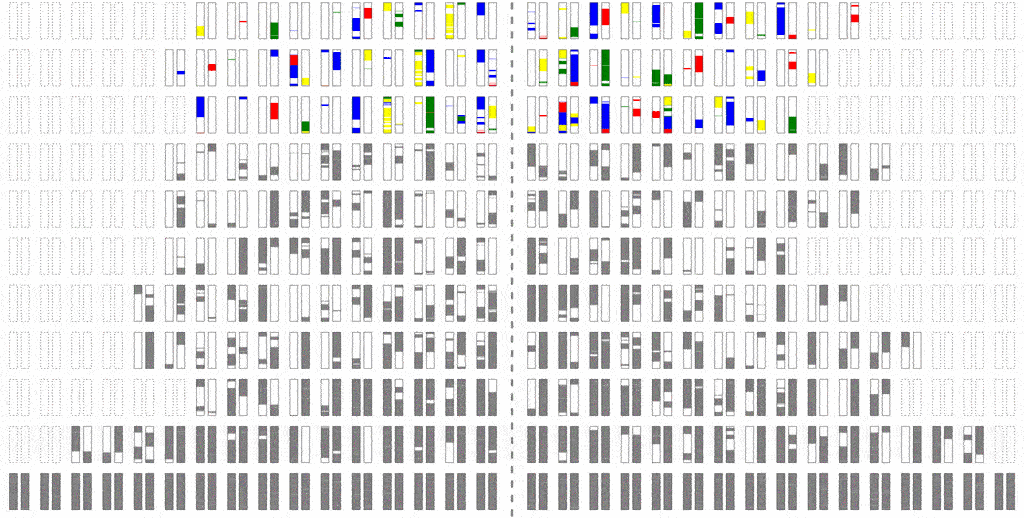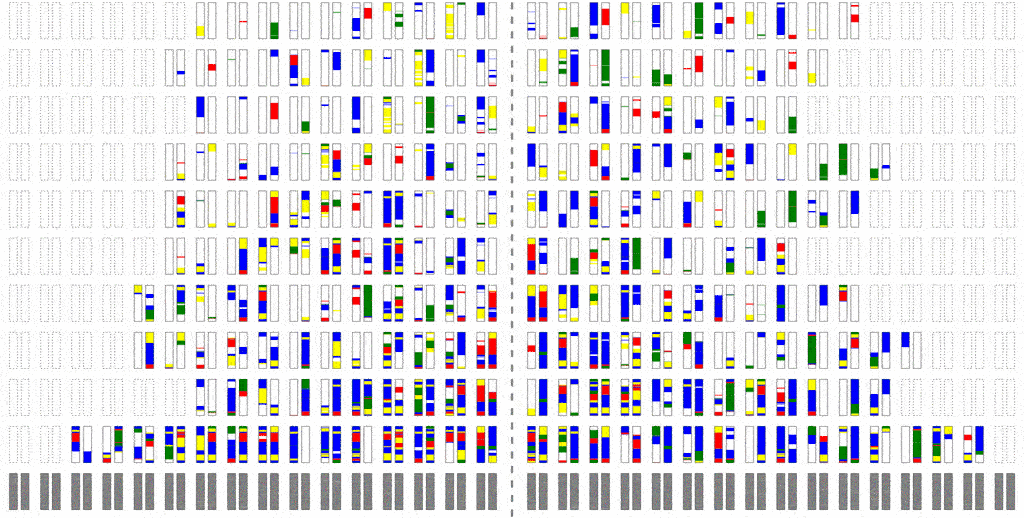
Look at how much more sparse this computation is, though it computes the same final result. The further back in time we go, the more sparse it becomes, allowing us to simulate further and further back with ease.
Even though an individual is ancestral, that doesn’t necessarily mean much of his/her genetic material is ancestral. To see why, consider that although you have two parents, you inherit only half of the DNA from each. From four grandparents, you only inherit about a quarter from each. From eight great-grandparents, you only inherit about an eighth. It doesn’t take very many generations before you are inheriting a smattering of genetic material from a wide range of different ancestors. In fact, it turns out that many of your genealogical ancestors will have passed on zero genetic information to you.
